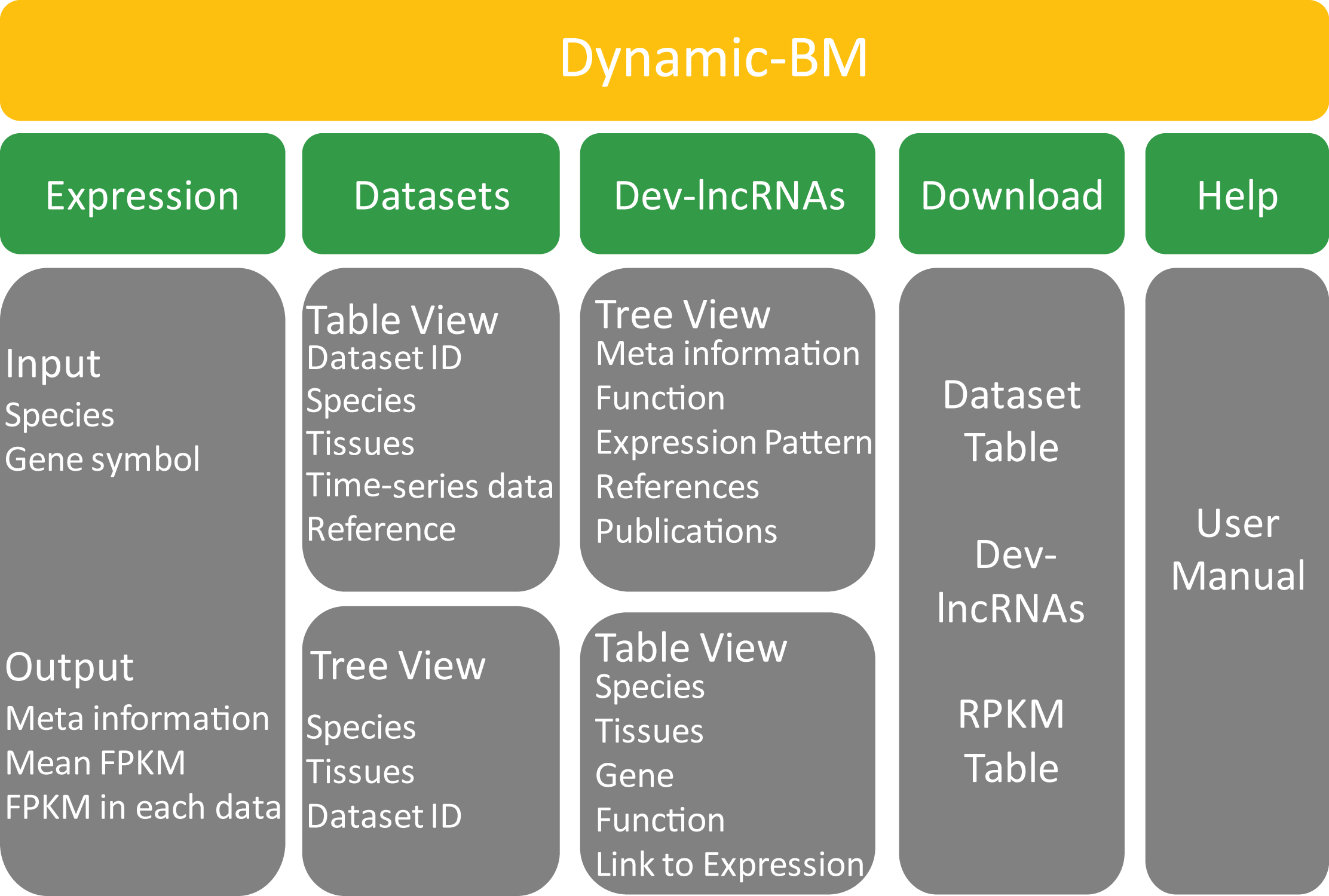
Dynamic-BM stands for Dynamic BodyMap, which provides massive and comprehensive dynamic RNA-seq data archives and analyses. The current version of Dynamic-BM contains 2,203 RNA-seq samples (114 datasets) of more than 20 tissues in 5 species.
The main sections of this website are Home, Expression, Datasets, Dev-lncRNAs, Analysis and Download. The Navigation bar on the right provides a quick access to the help file of each section.
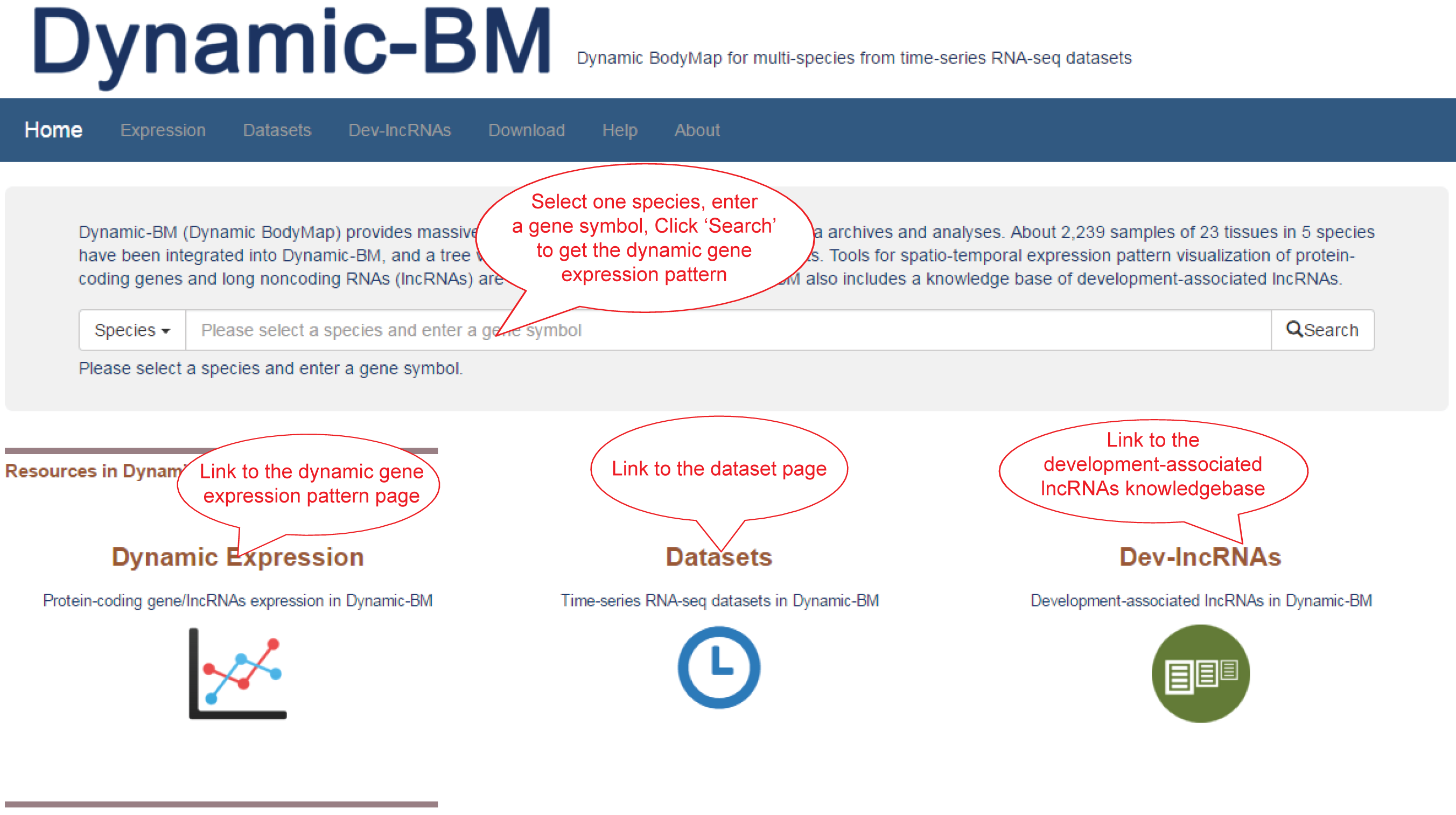
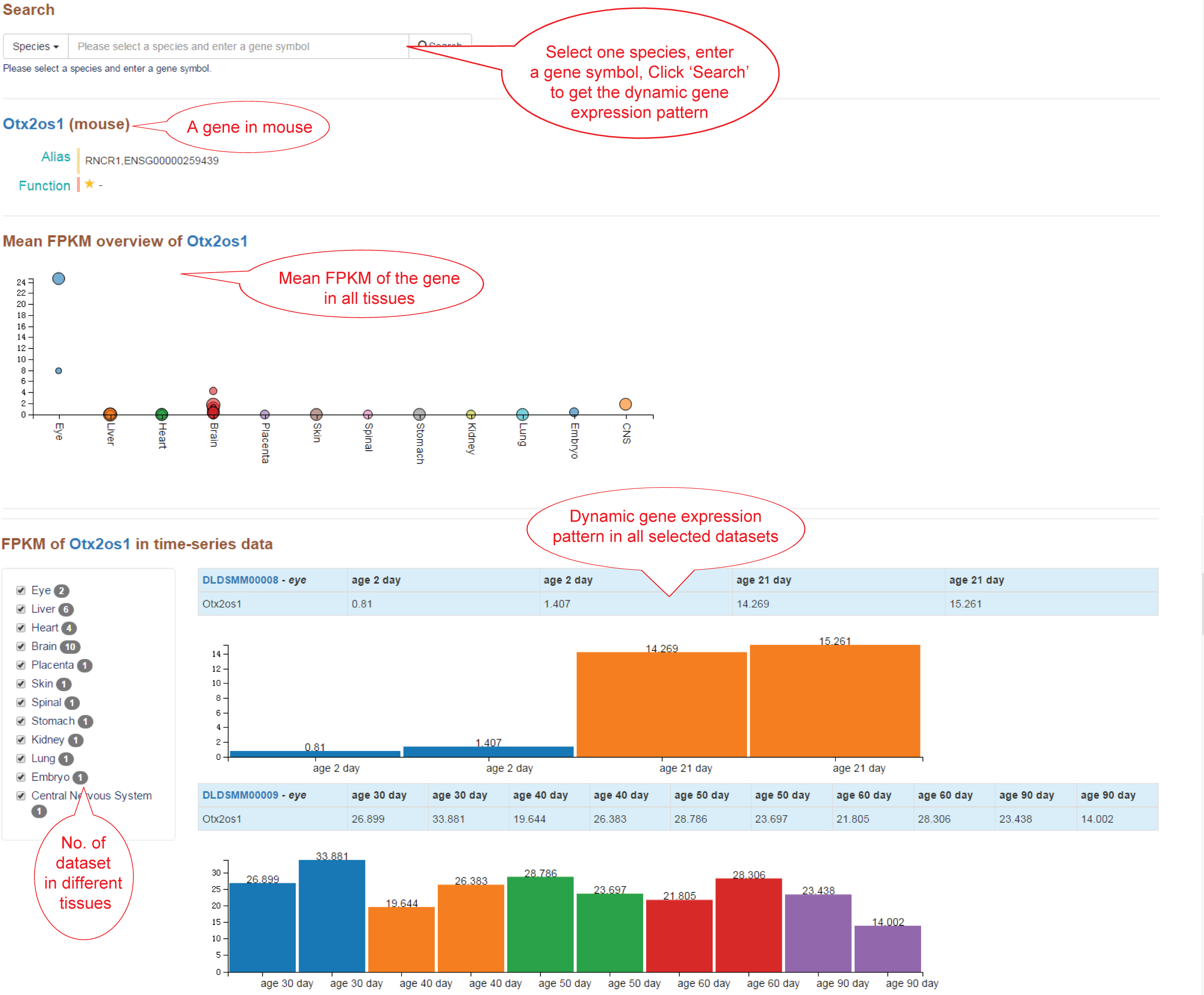
The Expression page provides: (1) Search box. select a species and enter a gene symbol, click 'Search' to get the dynamic gene expression profile. (2) Gene meta information. (3) Mean FPKM of the gene in all tissues. (4) Dynamic gene expression profile in all selected datasets.
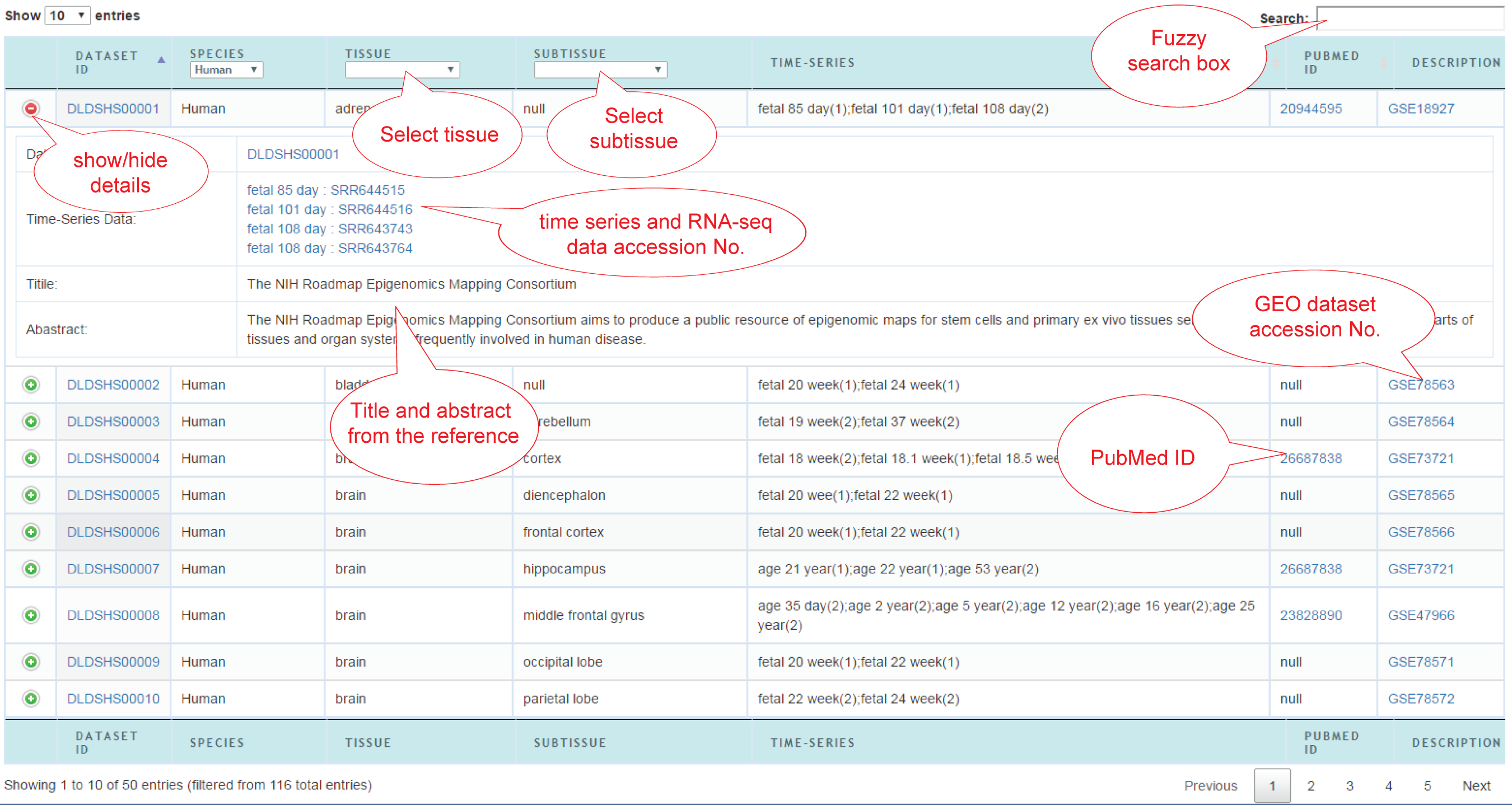
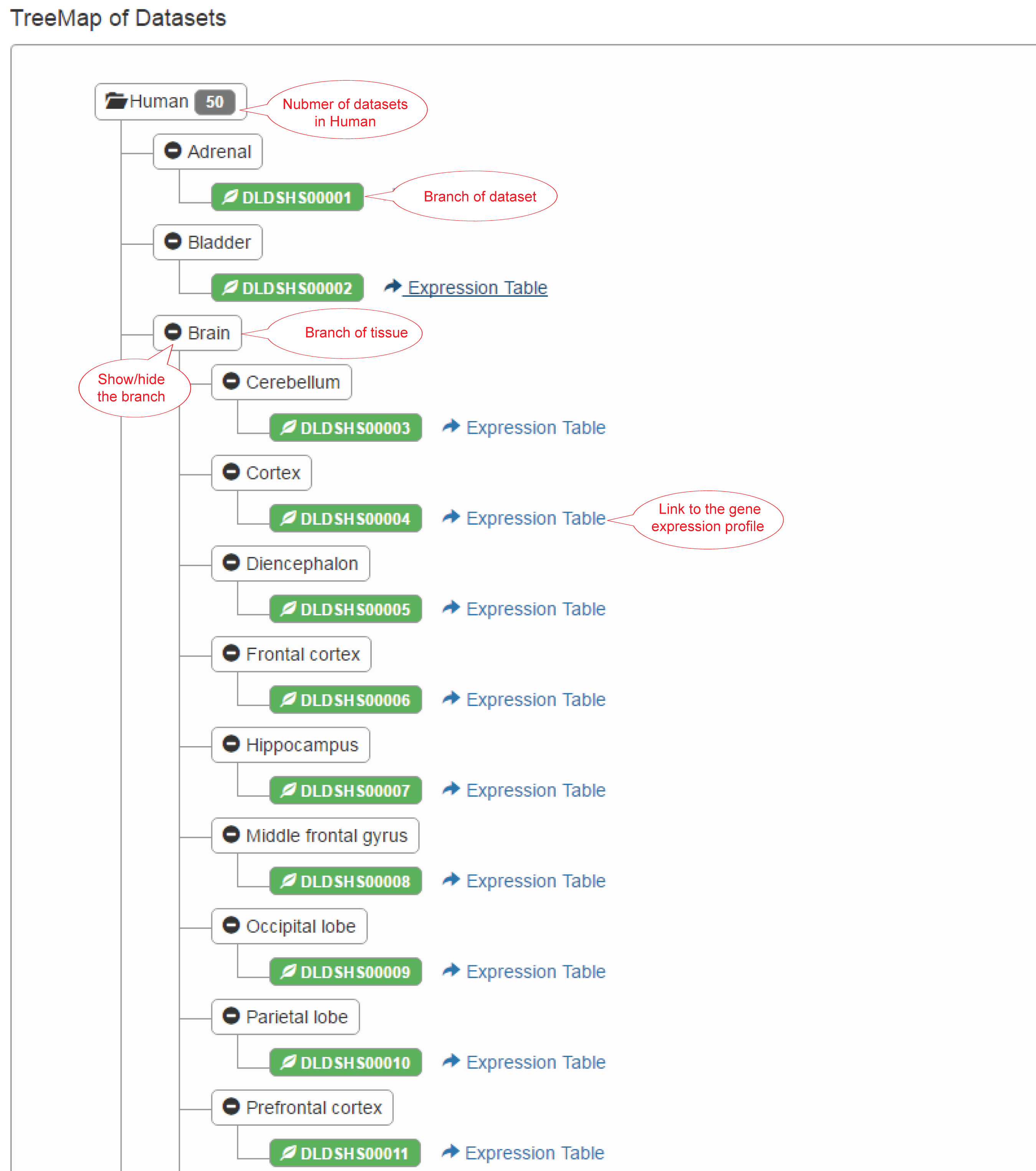
This page provides two ways to browse the datasets: (1) Table view; (2) Tree view.
(1) Table view: All the dynamic RNA-seq datasets are displayed in the table. Click '+/-' on the left of each row to show/hide the detailed information. Users can select species, tissue and subtissue to specify their interesting datasets. Click the accession number of dataset to jump to the gene expression profile.
(2) Tree view: All the dynamic RNA-seq datasets are organized in a tree. Take human as an example, the root of the tree is the species and the number of datasets. The branches are tissues and RNA-seq datasets from the tissues.
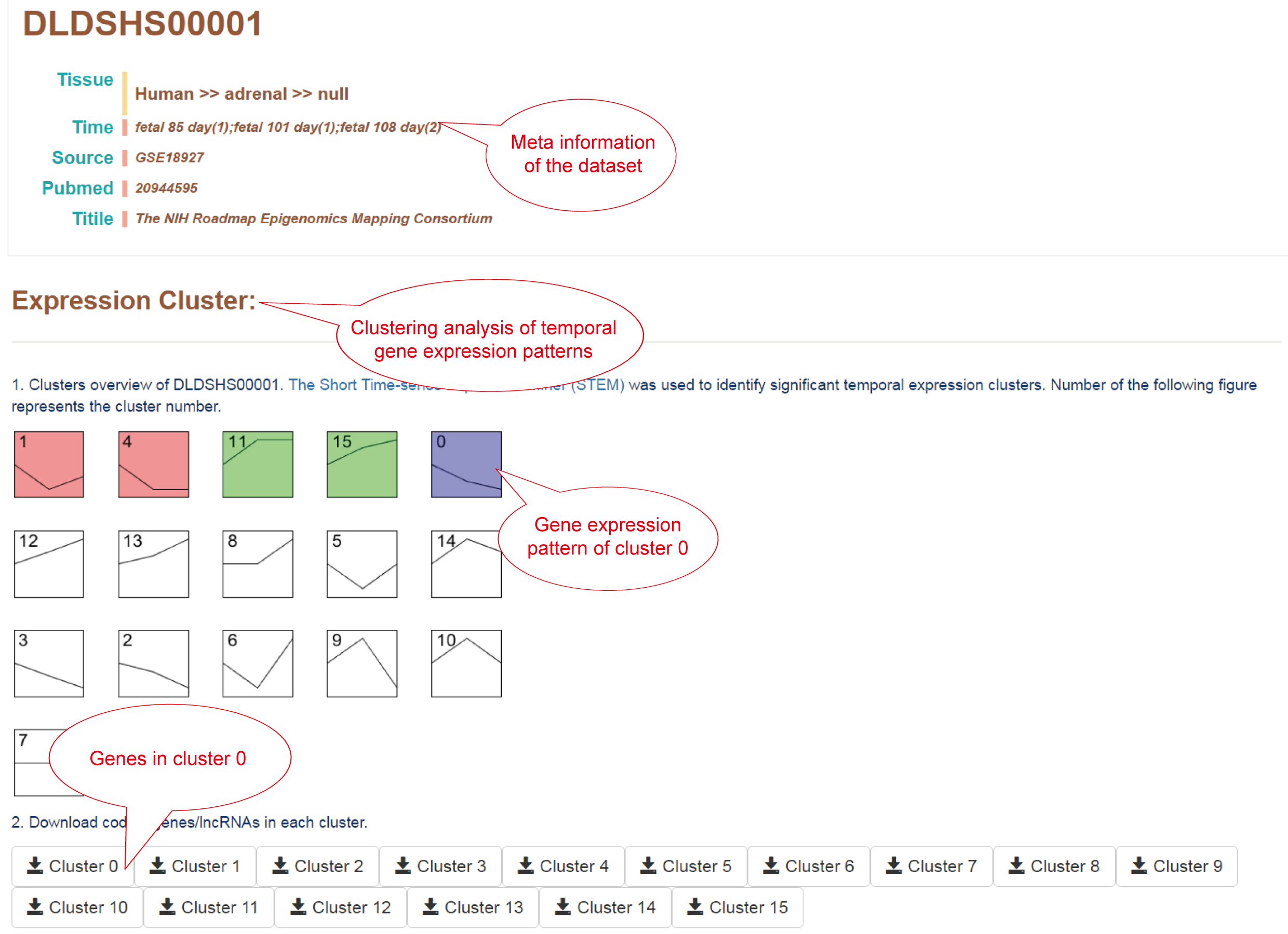
Clustering analysis of temporal RNA-seq datasets using The Short Time-series Expression Miner (STEM).
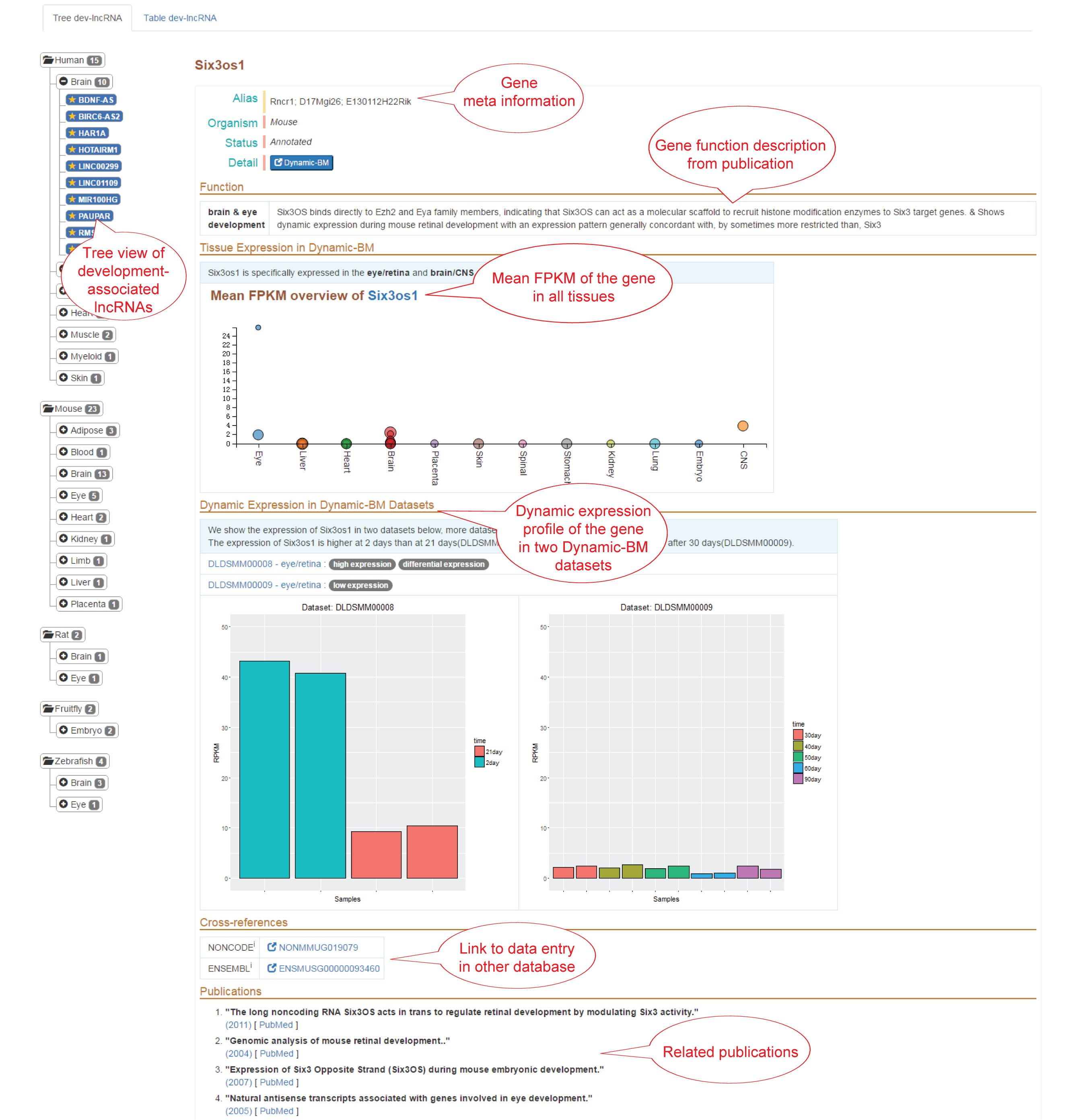
Dev-lncRNAs section is a knowledge base of development-associated lncRNAs. This page provides two ways to display these lncRNAs: (1) tree view; (2) Table view.
(1) Tree view: All the lncRNAs are organized in a tree. Take human as an example, the root of the tree is the species and the number of lncRNAs. The branches are tissues and lncRNAs from the tissues.Click '+/-' on the left of each branch to show/hide the detailed branches.
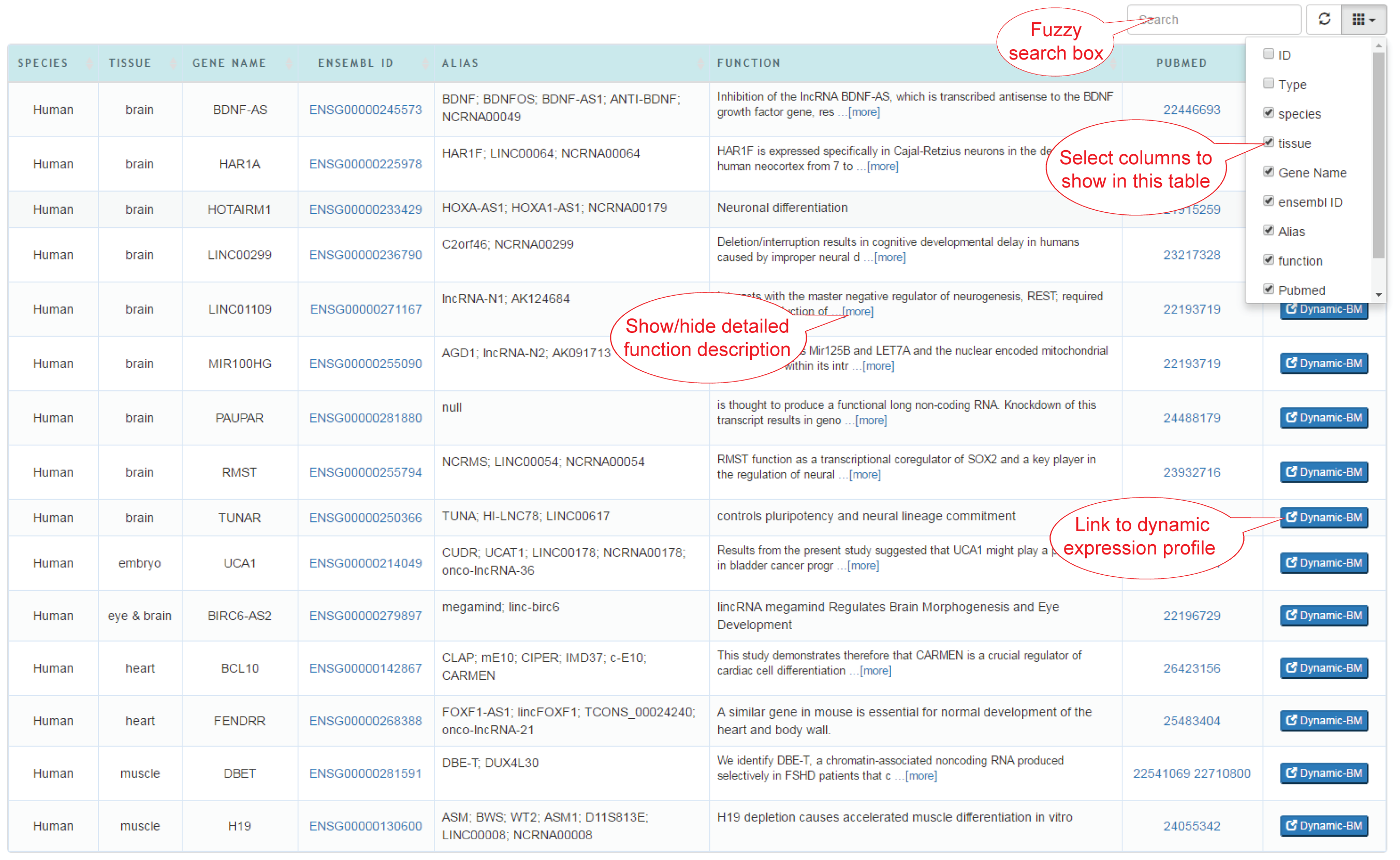
(2) Table view: All the lncRNAs are displayed in the table. Each row shows the meta information of each lncRNAs. Click 'Dynamic-BM' on the right of each row to jump to the dynamic gene expression profile of this lncRNAs.
Description of the time point of the datasets
We recorded the time point of each RNA-seq data in a uniform format. The keywords of the time points is described blow:
1. age: time after birth.
e.g. ¡°age 3 year¡± means 3 years after birth.
2. fetal: embryo development time.
e.g. ¡°fetal 22 week¡± means the embryo has grown for 22 weeks.
3. hpf/dpf:hours/days post-fertilization.
e.g. ¡°24 hpf¡± means 24 hours after fertilization.
4. prenatal: time before birth.
e.g. ¡°prenatal 2 day¡± means 2 days before birth .
We kept the original description of time points in some special cases, such as: ¡°0-2 hr after egg laying¡±, ¡°eclosion + 1 day¡±, ¡°embryogenesis: 2-4 cell stage¡± etc. Detailed description and overall experiment design can be obtained by clicking the link to the raw data.
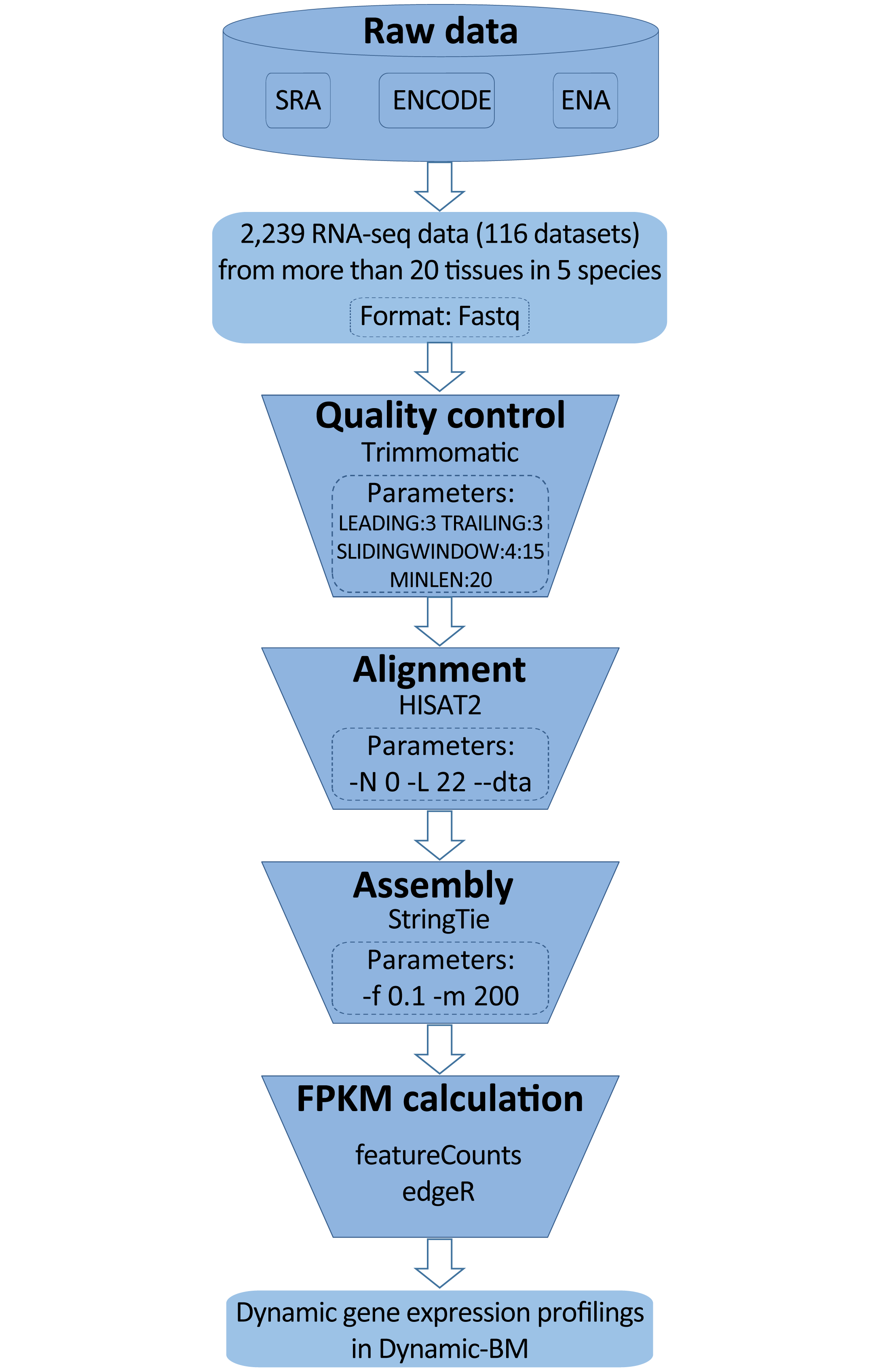
The software used in above pipeline are: (1) SRAToolkit v2.5.7; (2) Trimmomatic-0.35; (3) HISAT2-2.0.1-beta; (4) StringTie v1.2.2; (5) R-3.2.3; (6) edgeR 3.14.0; (7) BEDTools v2.19.1. And the reference genome for alignment in species are listed as follows:
If you have any questions or comments, please contact us.
Ya Cui
cui_ya(AT)163.com
Key Laboratory of RNA Biology, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China
Xiaowei Chen
chenxiaowei(AT)moon.ibp.ac.cn
Key Laboratory of RNA Biology, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China
Yiwei Niu
niuyiwei(AT)moon.ibp.ac.cn
Key Laboratory of RNA Biology, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China